Steve Matheson has a great post today on How to Evolve a New Protein in (about) 8 easy steps, worth quoting in detail:
How, exactly, does a protein acquire a new function during evolution? This is one of those “big questions” in evolutionary biology. Broad concepts such as gene duplication are quite helpful in formulating explanations, but the specific question raised is focused on the details — the actual steps — that must occur during the step-by-step modification of a protein such that it performs a different job than the proteins from which it has descended. The constraints on the process of change are significant, and the issues are similar to those I discussed when describing the concept of fitness landscapes in morphospace. The problem, basically, is this: how can you change a protein without wrecking it in the process? In other words, can you get from function A to function B, step by step, without passing through an intermediate form, call it protein C, which is worthless (or even harmful)?
These are precisely the questions addressed in an elegant set of experiments reported in two reports over the last year or so. The second article, by Ortlund et al., was reported in the 14 September issue of Science, and built on work reported in Science in April 2006. Their studies focused on two closely-related proteins that are receptors for steroid hormones. In this case, the steroids of interest are corticosteroids (the kind often used to treat inflammation; think cortisone) and a mineralocorticoid (a less well-known hormone, aldosterone, that regulates fluid and salt intake). The hormones are structurally similar (being steroids).
Joseph Thornton, at the University of Oregon, has been studying the origins of these receptors for about 10 years, and has assembled an interesting (and detailed) account of their history. The basic outline is as follows: the original steroid receptor was an estrogen receptor, and is extremely ancient, apparently arising “before the origin of bilaterally symmetric animals” (Thornton et al., Science 2003). (That’s seriously ancient, sometime in the Cambrian or earlier.) The progesterone receptor seems to have arisen next, followed by the androgen (i.e., testosterone) receptor. (Now that’s intriguing.) Fairly late in this game, the two receptors of interest to us here, the corticosteroid receptor and the mineralocorticoid receptor, were added to the vertebrate repertoire. The two modern receptors are thought to descend from an ancestral corticosteroid receptor, which underwent a gene duplication. Hereafter, I’ll refer to the receptors as the corticosteroid receptor and the aldosterone receptor, hoping that all the jargon won’t obscure the message.
In a widely-discussed paper published in Science a year ago (Bridgham et al., Science 2006), Thornton’s group determined the most likely DNA sequence of this ancestral gene, then “resurrected” it, meaning simply that they created that very DNA sequence in the lab. (Determining the ancestral sequence was a nifty piece of work; actually making the DNA is quite straightforward, especially if you have a little dough.)
Their experiments showed that the ancestral receptor could bind to a hormone that didn’t exist yet (aldosterone) while it was functioning as a receptor for corticosteroids. In other words, the receptor was available for activation by aldosterone long before aldosterone was around. (All jawed vertebrates make corticosteroids, but only tetrapods make and use aldosterone, an innovation that occurred at least 50 million years later.) The modern corticosteroid receptor has since lost its ability to interact with aldosterone, and Bridgham et al. chart the most likely evolutionary path, at the molecular level, by which we and other tetrapods came to have a corticosteroid receptor that won’t bind to aldosterone. The surprising result, however, is the fact that the ancient receptor was able to bind aldosterone, millions of years before aldosterone is thought to have been present.
It gets even more interesting.
The most recent paper adds significantly to the picture, and introduces some genetic concepts that Behe’s fans should pray he understands. The authors (Ortlund et al.) took their analysis to a far more detailed level, by extending their previous observations in two ways. First, they assembled a detailed family tree for the receptors, by looking at DNA sequences from living species known to represent various branches on the tree of life. In other words, they chose organisms such as lampreys, bony fish, amphibians and mammals, and examined their DNA codes (for the receptors) to find the changes that occurred in each branch of the lineage. Now, please stop and think about this, because it’s really cool. What the authors did was mine existing databases of DNA sequence data, pulling out the sequences of the two types of receptors from 30 different vertebrate species. You could repeat this part of the experiment right now, by referring to their list of organisms in Supplemental Table S5, which provides the ID codes needed to locate the DNA sequences in the Entrez Gene database. Then they charted the changes in the DNA sequence in the context of the tree of life as sketched out in the fossil record. The tree they assembled includes all the steroid receptors, and I’ve annotated it a little if you want to have a look. They used this tree to guide their further experiments, as I’ll explain below. The second thing they added to their previous analysis was an analysis of the 3-D structure of the various postulated intermediates in the evolutionary pathway. And they accomplished this by making proteins from the “resurrected” genes, then crystallizing them and using X-ray diffraction techniques to determine their precise structures.
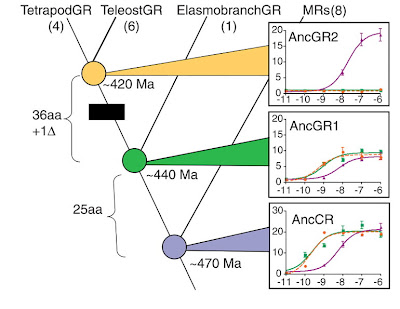
Examination of their receptor family tree revealed something interesting. Most vertebrates have highly specific receptors: the corticosteroid receptor isn’t strongly stimulated by aldosterone, and vice versa. But some living vertebrates (skates, in particular) show a different pattern: the corticosteroid receptor isn’t all that specific for cortisone. Because the ancestral receptor also lacked specificity (as shown in the 2006 paper), the authors concluded that the receptor acquired its discriminating taste at some point between the branching-off of skates (and their kin) and the separation of fish from tetrapods. Their Figure 1 is a little crowded, but it illustrates this nicely:
To follow the evolutionary narrative in this graph, start at the blue circle, which represents the ancestral receptor that was “resurrected” in the 2006 paper and that happily binds to both corticosteroids and aldosterone. (The graphs on the right side of the figure demonstrate the specificity, or lack thereof, of the receptors at different times in history.) There’s a branch leading up and to the left, to the various GRs (corticosteroid receptors), and one leading up and to the right, to the MRs (aldosterone receptors). At the green circle, another branching event occurred, 440 million years ago, at which point certain groups of fishes (skates among them) branched off, up and to the right. The receptor at that point is an ancestral corticosteroid receptor, and it still isn’t specific for corticosteroids. But the receptor at the yellow circle, in the common ancestor of tetrapods and bony fishes, is specific. The authors conclude that specificity arose between those two points, between 420 and 440 million years ago. With some (deliberate?) irony, they indicate that process with a black box.
This will definitely be worth watching over the next few years.